
Build Clusters Based on PTM Data Under Like Conditions
MakeClusterList.RdThis function takes the post-translational modification table and runs it through three calculations of distance: Euclidean Distance, Spearman Dissimilarity (1 - |Spearman Correlation|), and the average of the two of these. These calculations find the 'distance' between ptms based upon under what conditions they occur. These matricies are then run through t-SNE in order to put them into a 3-dimensional space. Additionally, the intersection of these 3 clusters is also created. A correlation table is also produced based on the Spearman Correlation table.
Arguments
- ptmtable
A dataset for post-translational modifications. Formatted with row's names containing PTM names. The column names should be drugs. Values are numeric values that represent how much the PTM has reacted to the drug.
- keeplength
Only keep clusters of ptms whose size is larger than this parameter. (I.e keeplength = 2 then keep ("AARS", "ARMS", "AGRS") but not ("AARS", "ARMS")); default is 2
- toolong
A numeric threshold for cluster separation, defaults to 3.5.
Value
A list with these data structures at the given index:
1 (Consensus Clusters as a list): Clusters in all 3 distance metrices as a list.
2 (Consensus Clusters as an adjacent matrix) A matrix containing values of 0s and 1s depending on if the PTMs are cocluster with other PTMs, rows and columns are unamed.
3 (PTM Correlation Matrix) A data frame showing the correlation between ptms (as the rows and the columns) with NAs placed along the diagonal.
Details
Please note: t-SNE involves an element of randomness; in order to get the same results, set.seed(#) must be called.
Examples
Example_Output <- MakeClusterList(ex_tiny_ptm_table) #Run function
#> [1] "Starting correlation calculations and t-SNE."
#> [1] "2025-10-20 22:12:10 UTC"
#> [1] "This may take a few minutes for large data sets."
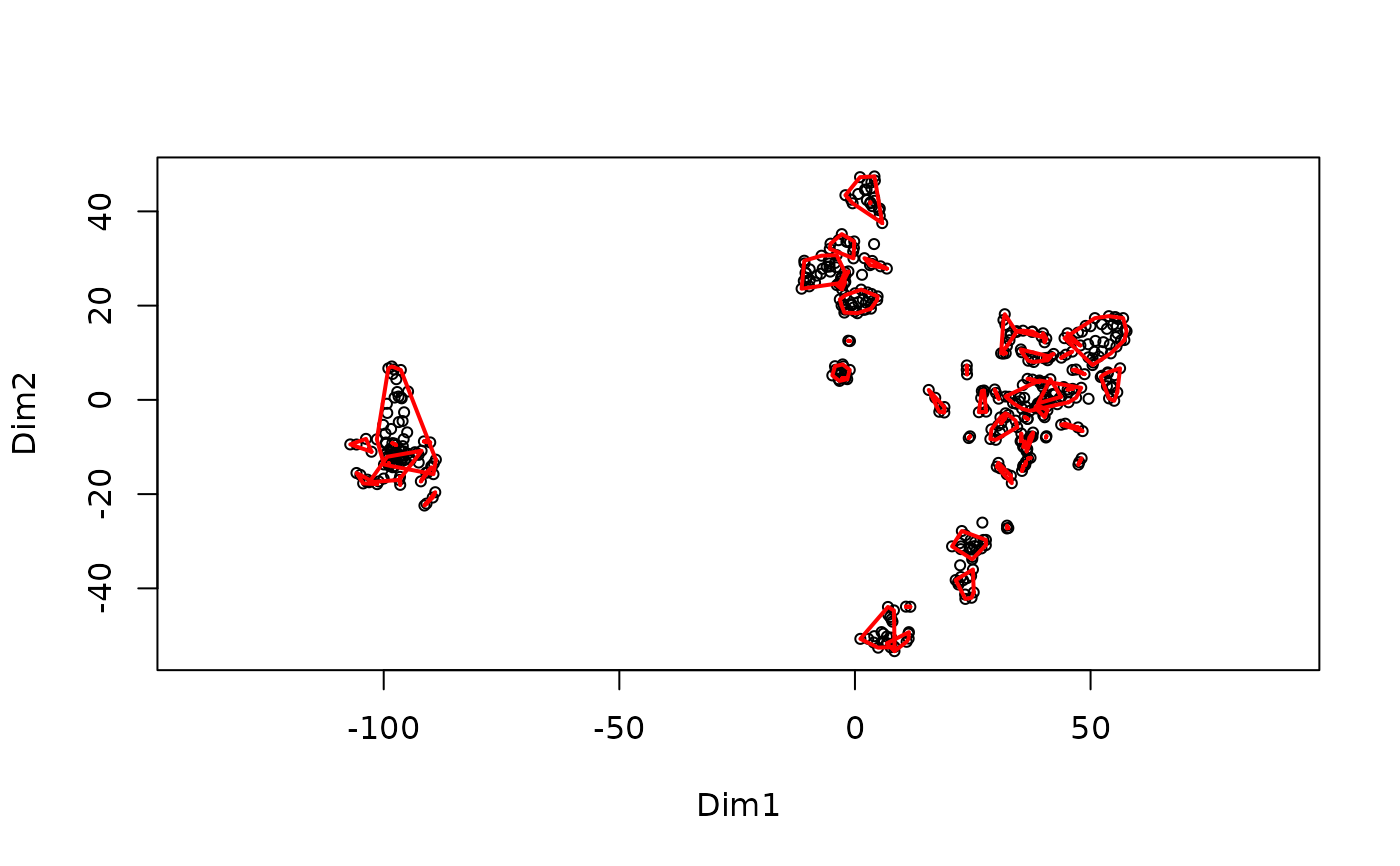
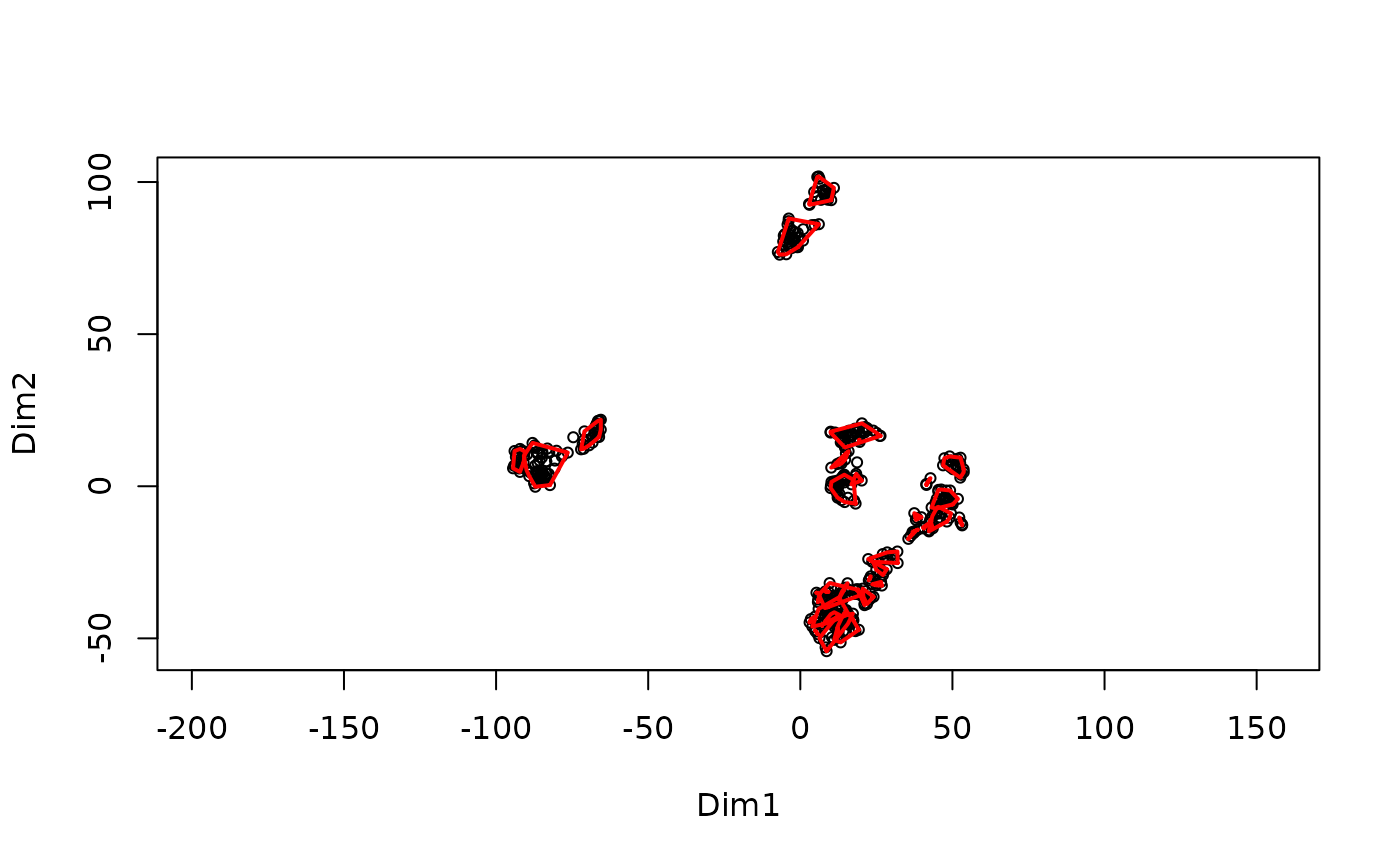
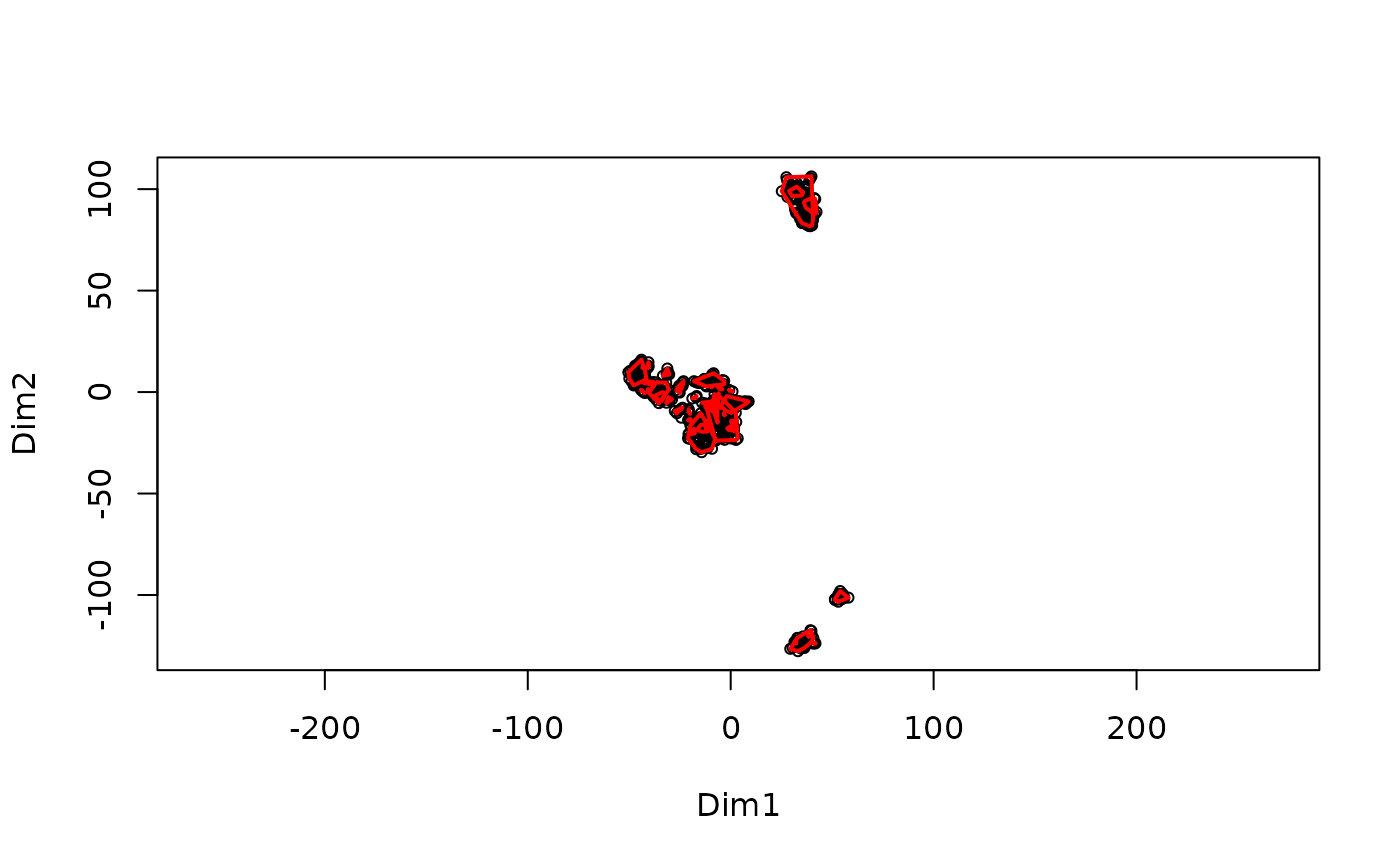 #> Starting FindCommonClusters at 2025-10-20 22:12:16.480787
#> [1] "2025-10-20 22:12:16 UTC"
#> [1] Total time: 0.0428409576416016
Example_Output[[1]][1:3] #Display data
#> $ConsensusCluster1
#> [1] "TNS3 p S602" "CEBPB p S141"
#>
#> $ConsensusCluster2
#> [1] "RACK1 p T229" "ATP6V1H p Y388" "IRS2 p Y632" "TNK2 p T829"
#>
#> $ConsensusCluster3
#> [1] "EPHA2 p Y588" "ATG101 p Y164" "SLC25A5 p Y195" "ALK p Y1584"
#>
#Do we want to have one for adj.consensus? Doesn't seem like it'd be very helpful to view.
utils::head(Example_Output[[3]][, c(1,2,3,4,5)]) #Display data
#> RNPS1 p Y205 PTPN6 p Y536 EIF2S1 p Y147
#> RNPS1 p Y205 NA 1 NA
#> PTPN6 p Y536 1 NA NA
#> EIF2S1 p Y147 NA NA NA
#> ACTG1 p T297; ACTB p T297 NA NA NA
#> RACK1 p T229 NA NA NA
#> EML4 p Y453 -1 -1 NA
#> ACTG1 p T297; ACTB p T297 RACK1 p T229
#> RNPS1 p Y205 NA NA
#> PTPN6 p Y536 NA NA
#> EIF2S1 p Y147 NA NA
#> ACTG1 p T297; ACTB p T297 NA NA
#> RACK1 p T229 NA NA
#> EML4 p Y453 NA NA
#> Starting FindCommonClusters at 2025-10-20 22:12:16.480787
#> [1] "2025-10-20 22:12:16 UTC"
#> [1] Total time: 0.0428409576416016
Example_Output[[1]][1:3] #Display data
#> $ConsensusCluster1
#> [1] "TNS3 p S602" "CEBPB p S141"
#>
#> $ConsensusCluster2
#> [1] "RACK1 p T229" "ATP6V1H p Y388" "IRS2 p Y632" "TNK2 p T829"
#>
#> $ConsensusCluster3
#> [1] "EPHA2 p Y588" "ATG101 p Y164" "SLC25A5 p Y195" "ALK p Y1584"
#>
#Do we want to have one for adj.consensus? Doesn't seem like it'd be very helpful to view.
utils::head(Example_Output[[3]][, c(1,2,3,4,5)]) #Display data
#> RNPS1 p Y205 PTPN6 p Y536 EIF2S1 p Y147
#> RNPS1 p Y205 NA 1 NA
#> PTPN6 p Y536 1 NA NA
#> EIF2S1 p Y147 NA NA NA
#> ACTG1 p T297; ACTB p T297 NA NA NA
#> RACK1 p T229 NA NA NA
#> EML4 p Y453 -1 -1 NA
#> ACTG1 p T297; ACTB p T297 RACK1 p T229
#> RNPS1 p Y205 NA NA
#> PTPN6 p Y536 NA NA
#> EIF2S1 p Y147 NA NA
#> ACTG1 p T297; ACTB p T297 NA NA
#> RACK1 p T229 NA NA
#> EML4 p Y453 NA NA