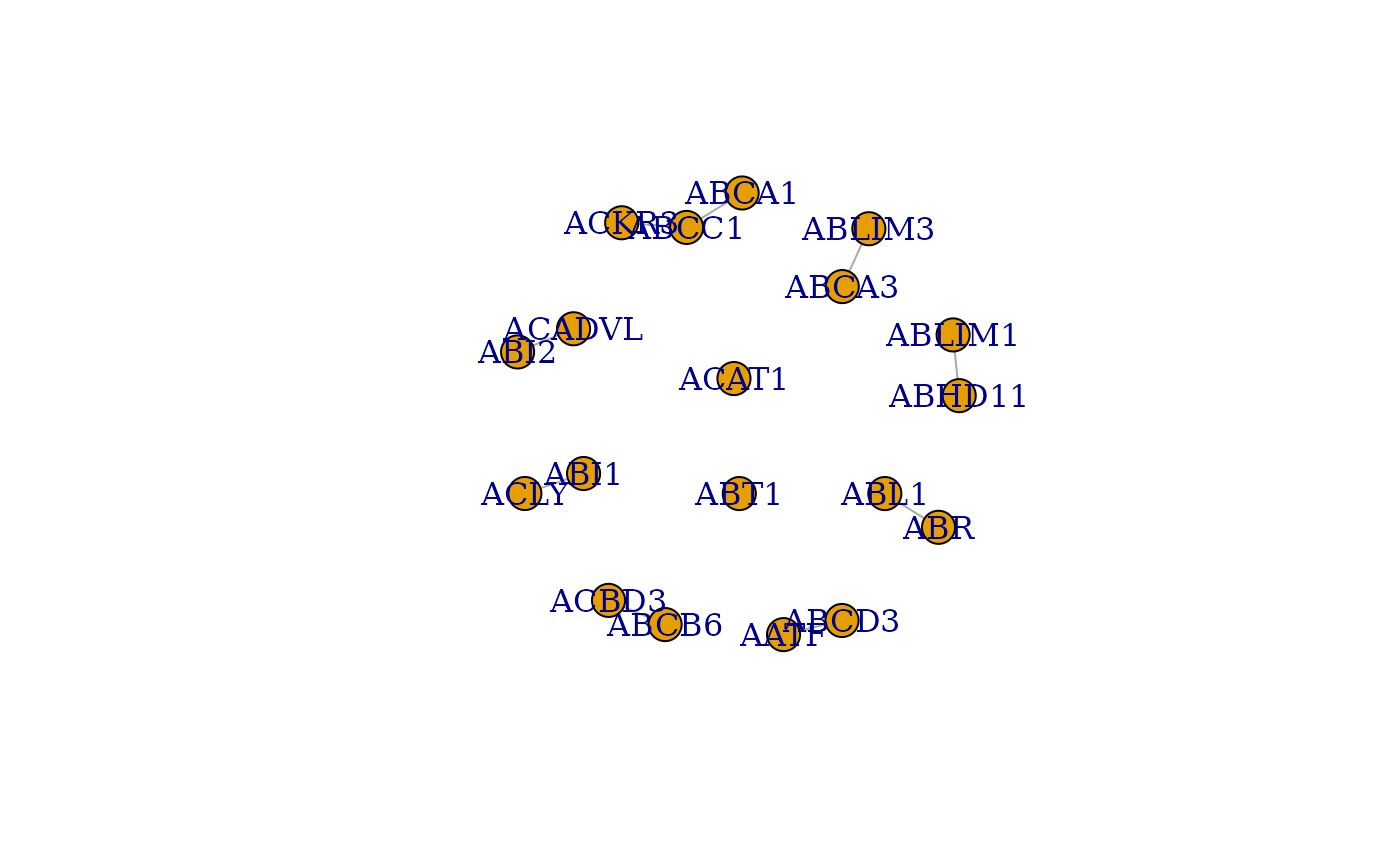
Make Correlation Network
MakeCorrelationNetwork.RdMake Correlation Network first finds the intersection between the Euclidian, Spearman, and SED cluster matrices in order to find the intersection between the three groups. It then adds the Genes in these PTMs to a list of common clusters and turns it into an adjacency matrix. This adjacency matrix is used to filter relevant data — clusters — from the Spearman correlation matrix. The resultant cocluster correlation network shows strength of relationships between proteins using the common clusters between the three distance metrics.
MakeCorrelationNetwork(
clusterlist,
ptm.correlation.matrix,
keeplength = 2,
clusters.name = "common.clusters",
cccn.name = "cccn.matrix"
)Arguments
- ptm.correlation.matrix
Correlation matrix made from ptm table
- keeplength
Only keep clusters of ptms whose size is larger than this parameter. (I.e keeplength = 2 then keep ("AARS", "ARMS", "AGRS") but not ("AARS", "ARMS"))
- cccn.name
The desired name for the output of the Correlation Network Matrix
- tsne.matrices
List containing matrices that contain Euclidean, Spearman, and SED t-SNE coords respectively
- lists.name
The desired name for the output of the list containing clusters of PTMs and Genes
Examples
MakeCorrelationNetwork(ex.clusters.list, ex.ptm.correlation.matrix, 1, "ex.common.clusters", "ex.cccn")